PSView - Help
This Web tool displays all PROSITE profile matches on all UniProtKB/Swiss-Prot entries with at least one match to a selected PROSITE profile.
Proteins are sorted and grouped by "architecture": PROSITE profile type alternation. For example proteins with a 'CUB, LDLRA_2, CUB, FZ' architecture are
proteins matched by (from N terminal to C terminal side) one or more CUB domains, then one or more LDLRA_2 domains and then again one or more CUB domains followed by one or more FZ domains.
Architectures are sorted by their complexity (number of profile type alternations), from complex ones to simpler ones. For each architecture only one 'template' protein is shown
(to view the other proteins use the expand button; proteins are sorted by their total number of matches).
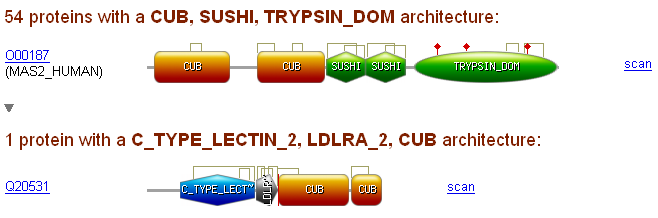
For each 'template' protein a graphical view in form of a downloadable png (Portable Network Graphics) image represent all its profile matches and detected features (see below).
Profile matches are represented as colored shapes with their PROSITE name. If a match overlaps with the previous one, it will be shown on a different line or if the overlap size is smaller
than 10% of the match size, the match will be shown on the same line, its overlapping start will be truncated and replaced by a vertical red bar (indicating that there is a small overlap).
If the mouse cursor is placed on a match, match information will be displayed in the browser status bar (protein name: domain name(domain AC) [match start - stop] (match score)) and
in a 'tooltip' box (domain name [match start - stop] (match score)).
For certain profiles, additional biologically meaningful information about residues inside matches is defined. This additional information comes from the mapping of biologically meaningful residues to PROSITE profiles. It is used to make functional/structural predictions of profile matches more accurate (as profiles show enhanced sensitivity over patterns, but because of their relaxed stringency loose functional/structural discriminativity).
If certain conditions expected for the functional and/or structural properties associated with the domain are fulfilled the properties are shown as 'Predicted features'. For each feature, the UniProtKB feature key, the position/range, the feature description (if any), and the condition that triggered the detection are shown.
Conditions can be specific amino acid inside hit, group of sub-conditions in which all conditions must be true in order for the group condition to be true, case between different sub-conditions/groups etc...
On the graphical view, features are shown on top of matches; depending on their type as bridges, horizontal 'bars', vertical pins.
For a protein of interest, if you want to obtain details about the detected features (Swiss-Prot feature key, position/range, description, and the condition that triggered the detection) and also about the absent features, use the link to the ScanProsite tool on the right of its image.

